|
My research consists of computer modeling of nanoscale and
biophysical systems. Because nanoscale objects are so tiny, it is very
difficult (sometimes impossible) to obtain a clear picture and
understanding of their structure and physical properties from
experimental measurements alone. Such limitations are not present in
the computer (virtual) world. I employ a computational technique called
molecular dynamics simulation along with 3D computer visualization to obtain
an atomic scale picture and understanding of nanoscale objects. Below are some
images and descriptions of a few projects that I have worked on. For more details, see my
publications. |
|
Structure of DNA-Carbon Nanotube Hybrids
|
|
|
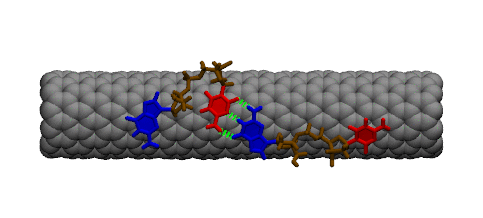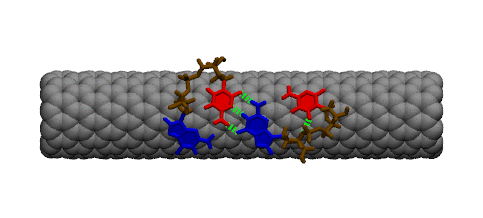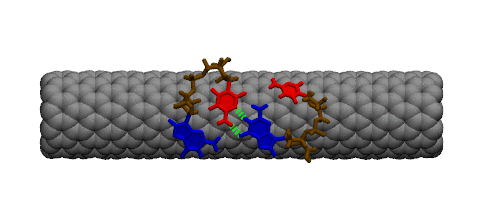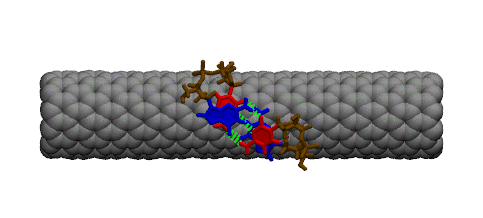
Even though DNA and carbon nanotubes have never encountered each other in nature, they are chemically compatible and can be combined to form hybrid materials that hold a wide range of useful properties. Current applications of DNA-carbon nanotube hybrids include carbon nanotube solubilization and sorting and chemical/biological sensing. Despite the importance of this material, its fundamental structure and physical properties are poorly understood. To gain insight into this nanomaterial, I have performed computer simulations to study the interactions between DNA and carbon nanotubes. Below are some images that illustrate some of my findings. |
|
|
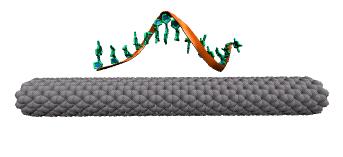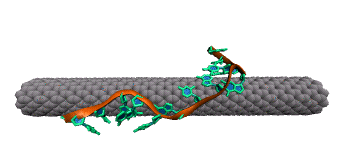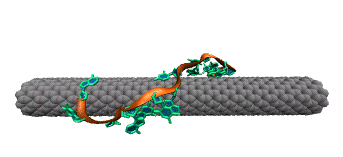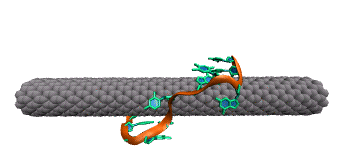
DNA attaches to a carbon nanotube via the π-π stacking interaction between the DNA bases (green rings) and the carbon nanotube sidewall. This interaction causes the DNA bases to stack on the surface of the nanotube.
|
|
|
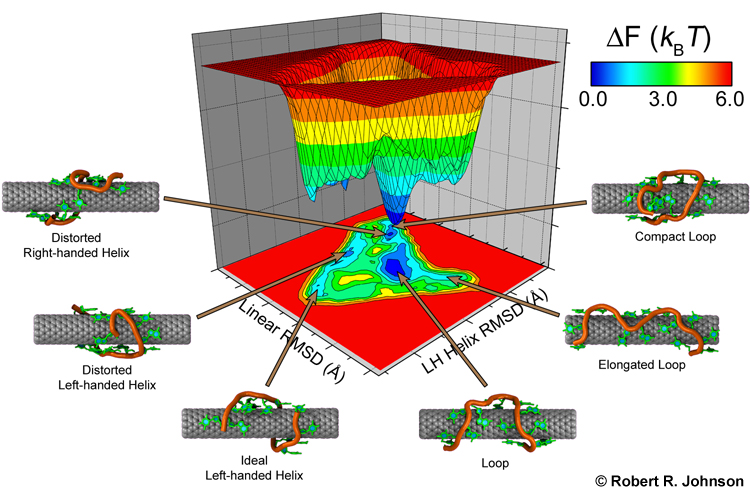
Single-stranded DNA is very flexible and can wrap around the nanotube in many ways. To identify the most favorable wrapping conformations A large-scale replica exchange molecular dynamics simulation was run on 2,048 CPUs for a month at the San Diego Supercomputer Center to compute the free energy landscape of a DNA-carbon nanotube hybrid. The results show that helical wrappings and U-shaped loop conformations are the most favorable.
Complementary DNA can hybdridize on the surface of carbon nanotubes. However, because of the strong interaction between DNA bases and the carbon nanotube, a large conformational change must occur within DNA prior to hybridization. The image below shows two single strands of DNA with sequence GpC. Two bases must lift off the surface of the nanotube to enable complete base pairing between the two strands.
|
|
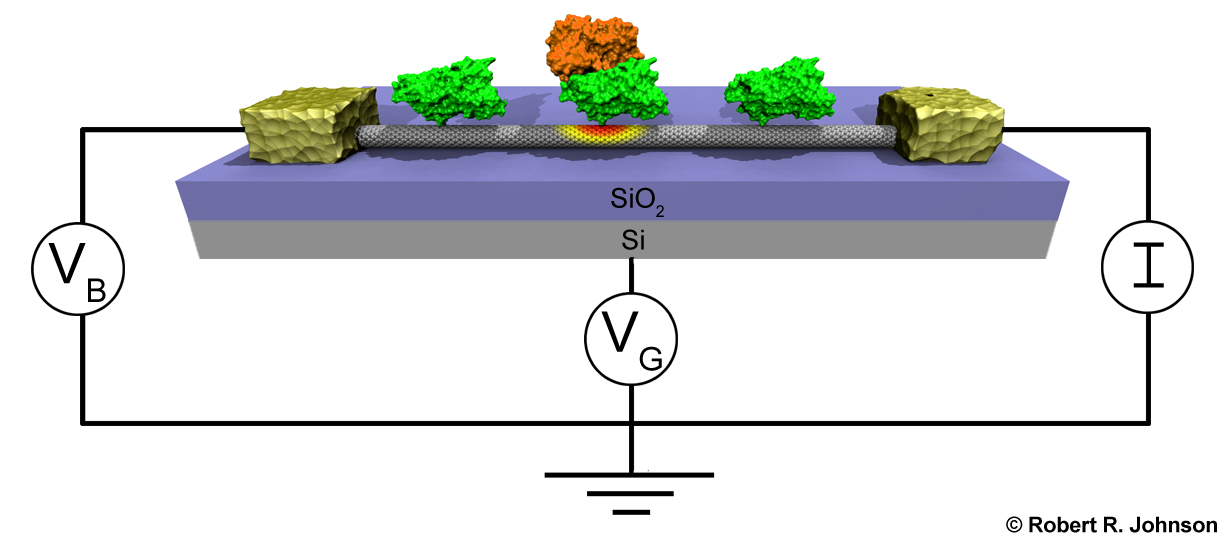
Simulation of Nano-Biosensors: Carbon Nanotube Functionalized with the Coxsackie-Adenovirus Receptor
|
|
|
Detection of viruses and other harmful biological agents has important applications in homeland security and medicine. Recently, Dr. Charlie Johnson's experimental nanoscience group at U. Penn has developed sensors capable of detecting proteins associated with the adenovirus (one of the viruses responsible for the common cold). These sensors are depicted below. They consist of the Coxsackie-adenovirus receptor protein (green) attached to a carbon nanotube (gray cylinder). Knob proteins (orange) that are located on the surface of the adenovirus bind to this receptor protein. This binding event changes the electrical properties of the carbon nanotube. Thus, by monitoring the electrical properties of the carbon nanotube, one can detect the presence of the adenovirus. I have used computer simulation to study how proteins interact with carbon nanotubes in order to help rationalize the fabrication and understanding of these sensing devices. |
|
|
|
|
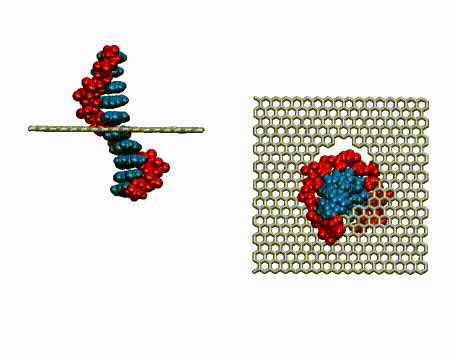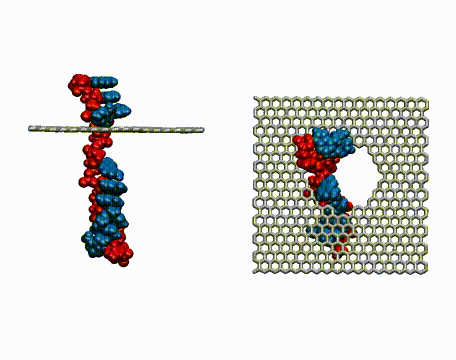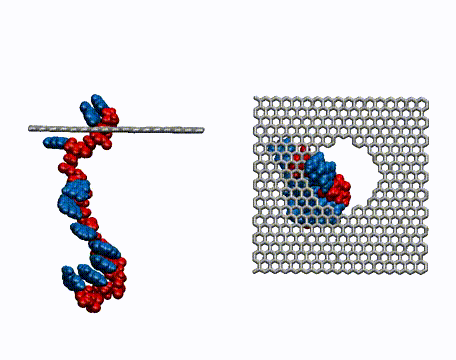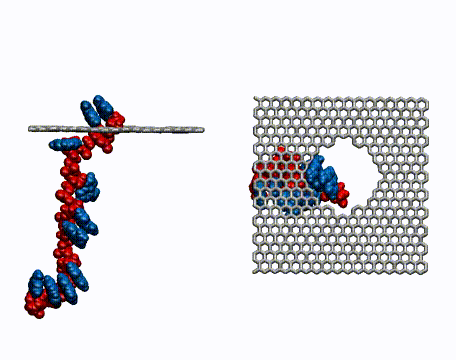
DNA Translocation through Graphene Nanopores
|
|
|
Determining the sequence of nucleotides that compose a DNA strand is of paramount importance in medicine and microbiology. Current techniques for DNA sequencing are complicated, error-prone, time consuming and only enable sequencing of short strands of DNA. Methods for the fast sequencing of genomic DNA are highly desired. This may be achieved in the near future by measuring the electrical properties of DNA bases as a DNA strand threads through nanoscale pores in graphene, an atomically thin, single layer of graphite. DNA carries a negative charge and is pulled through the nanopore by an electric field. Because of the strong interaction between DNA bases and graphene, the bases can be made to pass through the pore in single file. Ultrafast DNA sequencing technology attempts to read off the DNA bases as they travel through the nanopore. |
|
|
|
|


|
|